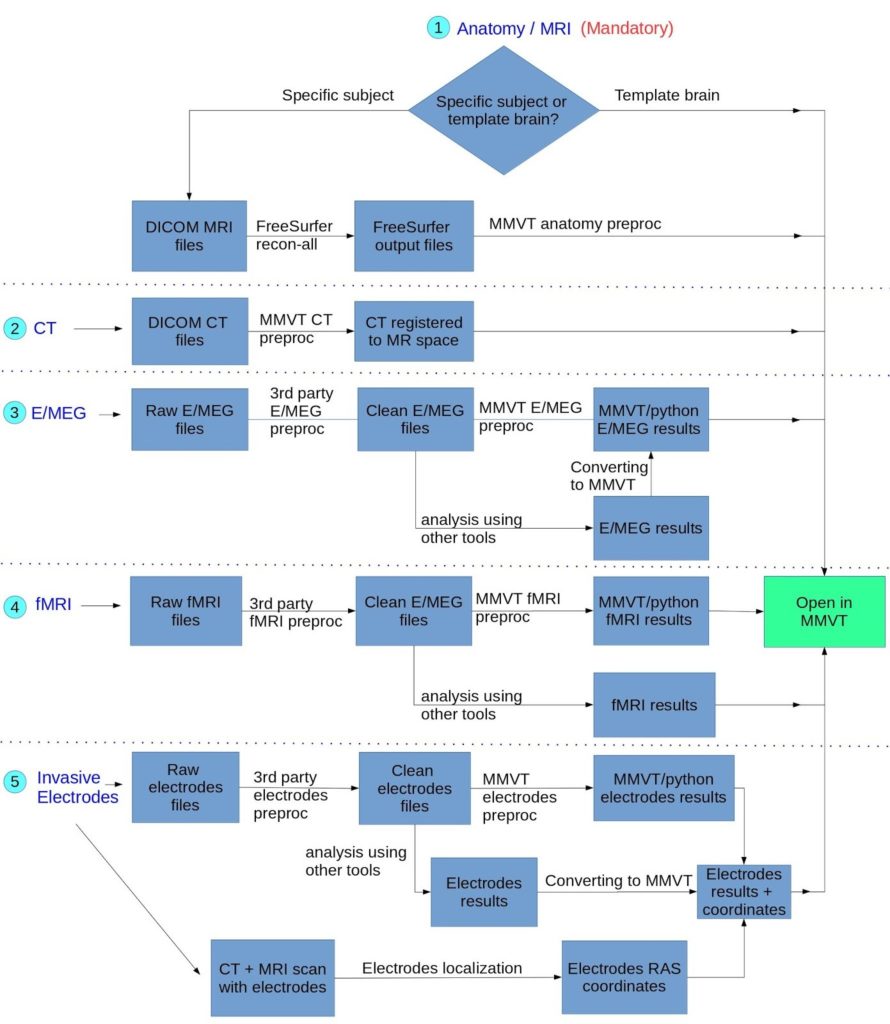
Preprocessing
- Reconstruction of the subject’s T1-MPRAGE using FreeSurfer recon-all module.
- Creation of a cortical atlas-specific for the subject, in case the atlas is not part of FreeSurfer (Desikan-Killiany, DKT, and Destrieux [1]). The atlas will be morphed from a template brain.
- Parcellating the pial and inflated surfaces to cortical labels given a cortical atlas. Each cortical label is being saved as a separate object that can be used later for visualization.
- Subcortical segmentation: The subcortical surfaces (based on the FreeSurfer volumetric atlas (aseg.mgz)) are generated using the marching cubes algorithm [15] and applying a Multidimensional Gaussian filter for smoothing.
- Creation of a spatial connectivity matrix between the vertices in the cortical surfaces. This matrix is being used for finding the neighboring vertices (a vertice’s neighbours are all the vertices connected to this vertice with an edge.). For example, the matrix is being used for calculating the contours of the cortical labels. This is done by finding all the vertices with neighbors from more than one cortical label.
- Creation of a morphing function between the pial and the inflated surfaces using Morph target animation [16]
- Importing objects into Blender. This creates the final Blender file that will include the cortical and subcortical structures, and the inflated surfaces.
- Creation of the dural surface. First, a volumetric file is being made using the FreeSurfer algorithm for quantifying local cortical gyrification [17]. Then, it’s converted to a surface using the marching cubes algorithm [15] and applying a Multidimensional Gaussian filter for smoothing. One possible usage of the dural surface is for “snapping” invasive electrodes on this surface by using Dykstra et al algorithm [18].
- Sub-segmentation of the cerebellum using the Buckner 2011 atlas [19]
- Creation of a flat map. This procedure is based on a FreeSurfer semi-automatic flattening procedure (mris_flatten). Unlike the Freesurfer procedure, where the user must cut the cortex manually, in MMVT this step is being done automatically, by using the contours of the DKT cortical atlas.
- Skull reconstruction: when T2 or CT scan is available, the skull inner and outer surfaces can be reconstructed using the marching cubes algorithm [15] or by using the surfaces of them Boundary Elements Model (BEM) were constructed using MNE [6] (mne_watershed_bem). After the reconstruction, the skull thickness is being calculated in the following way: From each vertice of the inner or outer skull surface, a ray is casted in the direction of the vertice’s normal until it hits the other surface [20].
For running the anatomy preprocessing, you’ll need FreeSurfer, which can be used only on a Linux or Mac OS. If you are using Windows, you’ll need to run this step on another machine, or on a virtual one. You can download an ubuntu virtual machine that already includes FreeSurfer here.
In case you do want to work on a specific subject’s brain, you’ll need to run the anatomy preprocessing script. Parts of the script are using FreeSurfer commands, so first you’ll need to download FreeSurfer, the dev version (look for “Development Version”). To source Freesurfer, you can:
Do it manually every time you open a new terminal: Set the directory where FreeSurfer was installed as an environmental variable typing:
export FREESURFER_HOME=/usr/local/freesurferThen source the setup script typing:
source $FREESURFER_HOME/SetUpbreeSurfer.shEdit the .bashrc (if you use a bash shell) and setup an alias Open the .bashrc file (or .cshrc, depends if you are using bash or csh) with a text editor:
gedit ~/.bashrcAdd these two lines
export FREESURFER_HOME=/usr/local/freesurferalias alias_name="source $FREESURFER_HOME/SetUpFreeSurfer.sh"Save the changes and start a new terminal. Every time you start a new terminal and type alias_name it will automatically source FreeSurfer.
cd inside the folder where the mmvt code is (/home/user/code/mmvt for example). Make sure your python is 3.4 and above. You can check that by running the following command:
python src/which_python.py
Part of the anatomy preprocessing is using Matlab (If you don’t have Matlab, we are really sorry, we’ll migrate this code to python ASAP. Meantime, you can contact us and we’ll run this step for you). Before running the script, you need to make sure you can run matlab by running “matlab” in the command line. If you areldy have matlab installed, but you can’t run it by calling “matlab” from the command line, you have two options:
- Copy the default_args.ini file from the resources folder, under the mmvt code folder. Edit the file and set your matlab_cmd full path command.
- On Linux based OS, add a matlab alias to the basrc/cshrc files. To make sure you can run matlab, run the following command:
python -m src.run_matlab
Now you can run the anatomy preprocessing. Here we are using ‘subj01’ as the name for the subject, you should replace it with your own subject’s name:
python -m src.preproc.anatomy -s subj01
The default parcellation atlas is the FreeSurfer aparc.DKTatlas40 atlas. If you want to use a different atlas, like the Lausanne250 for example, make sure your local fsaverage has the annotation files. You can download Lausanne250 and Lausanne500 files here.
python -m src.preproc.anatomy -s subj01 -a atlas_name
In the MMVT_DATA folder, you’ll find the atlas.csv file. Make sure the atlas you are using is there. In this file, you’ll find the aparc, aparc2009 and aparc.DKTatlas40 FreeSurfer atlases, and the Lausanne125, Lausanne250 and Lausanne500 atlases. The first column is the atlas friendly name (dkt for aparc.DKTatlas40 for example), the second column is the atlas real name, and the third is the atlas’s description.
If the FreeSurfer recon-all files aren’t placed locally, or you don’t want to write to this folder (in that case you should have chosen a different subject’s folder in the setup), you should add the –remote_subject_dir flag:
python -m src.preproc.anatomy -s subj01 -a atlas_name --remote_subject_dir /home/user/subjects/subj01
If you need to use ftp to reach the recon-all files, you should add the sftp, sftp_username and sftp_domain flags (you’ll be asked for the password in the command line):
python -m src.preproc.anatomy -s subj01 -a atlas_name --remote_subject_dir /home/user/subjects/subj01 --sftp 1 --sftp_username USERNAME --sftp_domain DOMAIN_NAME
You can also process multiple subjects at once by concatenating the subjects names with a comma (no spaces):
python -m src.preproc.anatomy -s subj01,subj02,subj03 -a atlas_name
Warning: If you wish to run the script on more than one subject, you shouldn’t run the script in parallel (in more than one terminal). This issue will be fixed in the future.
If you wish to get the recon-all files from a remote directory, for more than one subject you should add the {subject} template to the remote_subject_dir (the apostrophes are needed because of the ‘{‘ special character):
python -m src.preproc.anatomy -s subj01,subj02,subj03 -a atlas_name --remote_subject_dir "/home/user/subjects/{subject}"
Of Course, you can also use the sftp flags.
Troubleshoot: If for some reason the connection or the data transfer fails rerun the command by adding the overwrite_fs_files flag to overwrite the files:
python -m src.preproc.anatomy -s subj01 -a atlas_name --overwrite_fs_files 1
After this step is completed you are now ready to create the subject’s blend file (the file you’ll open in Blender):
python -m src.mmvt_addon.scripts.create_new_subject -s subj01 -a atlas_name
You can use the subjects flag instead of the s flag to create more than one Blend file:
python -m src.mmvt_addon.scripts.create_new_subject --subjects subj01,subj02,subj03 -a atlas_name
- Registration of the CT to the MRI using the FreeSurfer registration method, based on robust statistics to detect outliers and removes them from the registration, which leads to highly accurate registrations [21]. A mutual information error function is being used for cross modal registration.
- Creates an MRI and CT merged nifti file, to be plotted in MMVT slices viewer.
- Reading EEG and MEG from different caps and scanners and convert them into a Neuromag (fif) format.
- Artifacts removal, by using the Preconditioned ICA for Real Data (Picard) algorithm [22].
- Reads the sensors layout and creates a subject’s specific 3D mesh.
- Calculates the evoked response for a given task, to be plotted later on the MEG/EEG caps.
- Calculates the forward model, inverse operator [23], [24], and source localization per condition, by using the following inverse methods: minimum norm estimation (MNE) [25], dynamic statistical parametric mapping (dSPM) [26], standardized low-resolution brain electromagnetic tomography (sLORETA), exact low resolution tomography (eLORETA) [27], or beamformers like linearly constrained minimum variance (LCMV) [28], [29], recursively applied and projected multiple signal classification (RAP-MUSIC) [30], [31] , and dynamic imaging of coherent sources (DICS) [28], [29], [32], [33], [34]. The source localization can be computed for EEG, MEG, or by using both if EEG and MEG were stimulatingly recorded (MEEG).
- Computes the power spectrum density (PSD) by using the multi-taper method [35] on sensors space and cortical labels, given a cortical atlas.
- Finds functional regions of interest (ROIs) in a source localization solution, by using the surfaces connectivity matrix [7], [36].
- Calculates cortical labels and sensors frequency-domain and time-frequency-domain connectivity, based on estimates of the cross spectral densities and power spectral densities (CSD/PSD). The spectral densities can be estimated using a multi-taper method with digital prolate spheroidal sequence (DPSS) windows [37], [38], a discrete Fourier transform with Hanning windows [39], or a continuous wavelet transform using Morlet wavelets [40], [41]. The supported connectivity methods are: Coherence, Coherency, Imaginary coherence [42], Phase-Lock Value (PLV) [43], Pairwise Phase Consistency (PPC) [44], Phase Lag Index (PLI) [45], Unbiased estimator of squared PLI, Weighted Phase Lag Index (WPLI), and Debiased estimator of squared WPLI [46].
- Computes cortical labels space induced power in given frequency bands, using Morlet wavelets [40], [41].
For MEG resting state analysis, first, we need to create a time course per cortical label. This can be done using one call to the MEG preprocessing script:
python -m src.preproc.meg -s subject-name -a atlas-name -t rest -f rest_functions --l_freq low-freq --h_freq high-freq --windows_length window-length --windows_shift windows-shift
Where low-freq and high-freq are the band limit for filtering. Instead of reconstructing the source for all the raw data at once, the script divides the raw data into epochs (windows), and reconstruct the time course separately for each window. In case you are interested only in calculating a global measurement of the resting state data, you can set the windows-shift to be equal to the window-length (10s for example), and then just concatenate the windows. If you are interested also in calculating the resting state dynamics, set the windows-shift to a 1/n of the window-length, and calculate the connectivity measurement for each window. The consecutive steps are the following:
- Raw data (sensors level) filtering. On default, the raw data is also filtered to remove the power line noise using a notch filter (The default frequency is 60Hz. You can change it using the power_line_freq flag). If you don’t want to remove the power line noise, set the remove_power_line_noise flag to 0. The raw data can be filtered also by setting the flags low-freq and high-freq:
- low_freq < high_freq: band-pass filter
- low_freq > high_freq: band-stop filter
- only low_freq: high-pass filter
- only h_freq: low-pass filter
- The data is segmented into windows of window-length length (ms) and windows-shift (ms). On default, certain windows can be rejected according to the flags reject_grad, reject_mag, and reject_eog. If you prefer to take the risk and not to reject any of the windows, set the reject to 0.
- The forward model and inverse operator are calculated (if they don’t exist).
- The source is reconstructed for each window separately using the selected inverse method. The default is dSPM.
- For each label, according to the atlas-name (the default is aparc.DKTatlas40), the time course is extracted using the selected extract-mode (the default is mean_flip)
For example, you wish to calculate the resting state dynamics of 0.5s windows length in the alpha band, using laus125 as atlas, MNE as inverse method and calculate the pca-flip of each label:
python -m src.preproc.meg -s subject-name -a laus125 -t rest -f rest_functions -i MNE --l_freq 8 --h_freq 10 --windows_length 500 --windows_shift 100 --extract_mode mean_flip
The end result of this call is two files, labels_data_rh.npz, and labels_data_lh.npz. Each one contains two variables, data (numpy matrix, #cortical labels x T), and names, which is the cortical labels names. If you wish to run a resting state analysis, take a look here.
If you wish to call this script from python, take a look at the example in src.preproc.examples.meg, function calc_rest:
from src.preproc import meg
from src.utils import utils
args = meg.read_cmd_args(dict(
subject=args.subject,
mri_subject=args.mri_subject,
atlas='laus125',
function='rest_functions',
task='rest',
l_freq=3, h_freq=80,
windows_length=500,
windows_shift=100,
inverse_method='MNE',
))
call_main(args)In the following example, we’ll run the full MEG analysis pipeline. The data is task data (MSIT). It was cleaned using nTSSS (native temporal signal space separation). We’re planning to load the MEG and MRI data in the near future (morphed to a template brain).
Preprocessing
Most files will be read and created in the subject’s MEG folder /links/meg/task-name/subject (meg-folder). The files needed for MMVT will be created in the subject’s MMVT folder: /links/mmvt/mri-subject-name (mmvt-folder) The first step will be calculating the evoked responses in the sensors level. For that, you need to have two files in the meg-folder:
- Raw data file: meg-subject-name_msit_nTSSS-raw.fif
- Events file: meg-subject-name_msit_nTSSS_interference-eve.txt
Then, run the following command:
python -m src.preproc.meg -s meg-subject-name -m mri-subject-name -a laus250 -f calc_epochs,calc_evokes,read_sensors_layout -t MSIT --contrast interference --t_max 2 --t_min -0.5 --data_per_task True --read_events_from_file True --events_file_name {subject}_msit_nTSSS_interference-eve.txt --cleaning_method nTSSS
There is also an example written for MSIT task:
python -m src.preproc.examples.meg -s meg-subject-name -m mri-subject-name -f calc_msit -r calc_epochs,calc_evokes,read_sensors_layout
As a result, the following files will be created in the meg-folder:
- The raw info data: meg-subject-name_msit_nTSSS_interference-raw-info.pkl
- Epochs file: meg-subject-name_msit_nTSSS_interference-epo.fif
- Evokes file: meg-subject-name_msit_nTSSS_interference-ave.fif
And in the mmvt-folder/meg
- The evoked data in numpy format: meg_sensors_evoked_data.npy
- The evoked meta data (names, conditions and dt): meg_sensors_evoked_data_meta.npz
- The sensors positions (pos and names): meg_sensors_positions.npz
Importing The Data Into MMVT
To import the sensors evoked data, call the following command:
python -m src.mmvt_addon.scripts.import_meg_sensors -s mri-subject-name -a laus250
- Calculate the phase lag index (PLI) of the cortical labels
- Parcellated according to the Lausanne125 atlas
- Calculate, like in the fMRI, the coefficient of variation
- Find much faster dynamics, 0.5s windows length, and 0.1s windows shift
python -m src.preproc.fMRI -s subject-name -a laus125 -f calc_lables_connectivity --connectivity_modality meg connectivity_method=pli,cv --windows_length 500 --windows_shift 100
To call this function in code you should:
from src.preproc import connectivity as con
args = con.read_cmd_args(dict(
subject=args.subject,
atlas=’laus125′,
function=’calc_lables_connectivity’,
connectivity_modality=’meg’,
connectivity_method=’pli,cv’,
windows_length=500,
windows_shift=100
))
con.call_main(args)
- Morphing volumetric files from and to a template brain and between subjects, projection of a volumetric file on the cortical surface, and morphing surface files from and to a template brain and between subjects. The template brain can be determined automatically according to the vertices number in the surface files (these functions wrap the FreeSurfer commands vol2vol, mri_vol2surf, and mri_surf2surf).
- Average a contrast file over cortical labels given a cortical atlas and over the subcortical regions using a subcortical atlas. In the case where a time domain exists, it also calculates time series over cortical labels using a given cortical atlas, and over the subcortical regions using a subcortical atlas.
- Project volumetric file on the subcortical regions surfaces using the BallTree algorithm in scikit-learn [47], a machine-learning python package, and weighted sum over the distances of the sources to the surfaces.
- Finds clusters in a contrast file given the surfaces connectivity matrix [7], [36].
- Calculates the frequency-domain and time-frequency-domain cortical labels and subcortical regions connectivity, based on estimates of the cross spectral densities and power spectral densities (CSD/PSD), by using the same approach as in the EEG/MEG connectivity analysis.
If you wish to load FsFast results, the script will try to read the FsFast directories tree:
bold/{contrast_name}.sm05.{hemi}/{contrast}/sig.nii.gz
Also, the script assumes those directories are placed in the fMRI link directory, so the full directory tree should look as following:
links/fMRI/{task}/{subject-name}/bold/{contrast-name}.sm05.{hemi}/{contrast}/sig.nii.gz
You need to specify the task and the contrast name. You can also specify a contrast, or leave it empty to get all the contrasts:
python -m src.preproc.fMRI -s subject-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline
The atlas-name is needed for the clustering step. The algorithm will find cortical clusters and calculate their intersections with the labels, according to the selected atlas. In case the recon-all files are in a remote directory, like in the [anatomy preprocessing] (https://github.com/pelednoam/mmvt/wiki/Anatomy-preprocessing), you can set the remote_subject_dir:
python -m src.preproc.fMRI -s subject-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline --remote_subject_dir full-path-to-subject-recon-all-files
By default, the threshold for calculating the clusters is set to 2. If you wish to change it, you should use the threshold flag:
python -m src.preproc.fMRI -s subject-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline --threshold threshold-value
By default, the colors will be calculated symmetrically, from -abs(max(data)) to abs(max(data)). That makes sense if you loads fMRI contrast map. In case you want the color to be calculated from min(data) to max(data), you should use the symetric_colors flag:
python -m src.preproc.fMRI -s subject-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline --symetric_colors 0
If you wish to rerun the script for the same subject, and overwrite the previous results, you should use the overwrite_surf_data flag:
python -m src.preproc.fMRI -s subject-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline --overwrite_surf_data 1
If you wish to run the script on more than one subject, you should write the subjects names, separated with a comma, without spaces. If you also need to specify the remote directory, you should add {subject}:
python -m src.preproc.fMRI -s subject1-name,subject2-name,subject3-name -a atlas-name --contrast_name contrast-name -t task-name -f fmri_pipeline --remote_subject_dir full-path-to-subject-recon-all-files/{subject}"
Make sure you are using apostrophes in the remote folder name, the ‘{‘ character can be problematic.
In case you just want to load fMRI/PET volume file (nifty image), the preprocessing script will also project the volume data on the hemispheres:
python -m src.preproc.fMRI -s subject-name -a atlas-name
In this case, the volume file will be located in:
links/fMRI/subject-name/subject-name.{format}
Where format can be nii, nii.gz or mgz. In case you want to set the volume file name, you should use the volume_name flag:
python -m src.preproc.fMRI -s subject-name -a atlas-name --volume_name volume-name
Now the volume file will be located in
links/fMRI/subject-name/volume-name.{format}
In case the files are located under a task folder, like in the FsFast example, you should set the task flag:
python -m src.preproc.fMRI -s subject-name -a atlas-name --volume_name volume-name -t task-name
Now the volume file will be located in
links/fMRI/task-name/subject-name/volume-name.{format}
In case you wish to load PET data, you need to use the is_pet flag. Also, in PET raw data, you most probably don’t want the colors to be calculated symmetrically, and the threshold should be set to a meaningful value, possibly zero:
python -m src.preproc.fMRI -s subject-name -a atlas-name --volume_name volume-name -t task-name --threshold 0 --symetric_colors 0 --is_pet 1
First, you should call the clean_4d_data function:
python -m src.preproc.fMRI -s subject-name -f clean_4d_data --fmri_file_template=file-template
Where file-template is the usually a wildchar expression for finding the raw file in the subject’s fMRI folder. You can also create a link to the original file. For example, if your file’s name is f.nii, use can set the –fmri_file_template to be “f.*”.
This function is basically a wrapper for the following freesurfer calls: preproc-sess, tkregister-sess, fcseed-config, fcseed-sess and selxavg3-sess. Those calls require a functional subdirectory, which as default is ‘rest’, but you can change it using the –fsd flag. Also, the default values for fwhm, lfp and nskip are 6, 0.08 and 4 respectivly, and you can change them using the flags –fwhm, –lfp and –nskip.
Usually, these freesurfer functions are morphing the data into the template brain. For doing so, you need to set the –template_brain flag to a template brain, like fsaverage, or fsaverage5. If you are not setting this flag, the data will remain in the subject’s MRI space. This option is preferable if later you want to import the data into the subject’s MMVT file. The function’s output is two files, one for each hemisphere: ‘{}.sm{}.{}.{}.mgz’.format(fsd, int(fwhm), trg_subject, hemi). If you’ve picked the default values, didn’t morph the data to a template brain, and set fsd to rest for example (if the data is resting state fMRI), the files will be named rest.sm6.subject-name.rh.mgz and rest.sm6.subject-name.lh.mgz
Next, if you wish to extract the time series from the cortical labels, you should call the analyze_4d_data function:
python -m src.preproc.fMRI -s subject-name -a atlas-name -f analyze_4d_data --fmri_file_template "*.{hemi}.{format}"
You can set the fmri_file_template to be more specific like ‘restsm6{hemi}.mgz’ for example. The function will calculate the mean of each label for each time point. If you don’t want to calculate the mean for all the labels, you can set the –excluded_labels flag. The default value is ‘corpuscallosum,unknown’. If you wish to calculate a different measure than the mean, you can set the –labels_extract_mode flag, where the possible values are mean,pca and cv (coefficient of variation). You can use more than one (mean,cv for example).
If you have the recon-all files of your subject, you just need to use the same approach like in fMRI/PET raw data. In case you don’t have the subject’s MRI, you’ll need to morph the result in SPM to the SPM template brain. Then, use colin27 as your subject-name when calling to the fMRI preprocessing (you’ll need to download colin27 first from here:
python -m src.preproc.fMRI -s colin27 -a atlas-name
If you wish to load group averaging/comparison results, you first need to convert the SPM results to FreeSurfer format. In the SPM directory, you’ll find spmT_0001.img and spmT_0001.hdr. In the command prompt, you’ll need to run the following command (after sourcing FreeSurfer):
mri_convert spmT_0001.img results-name.nii
Where results-name can be the subject’s name in case of subject vs group comparison, or any other name you find suitable. After that, copy the nii file to colin27’s fMRI results folder, which can be (as described above) in links/fMRI/colin27 or in links/fMRI/task-name/colin27 if you specify the task (using the -t flag). The file should be copied to the colin27 folder because, for group averaging/comparison, SPM uses its template brain, which is colin27. After that, you can load this file as described in the loading fMRI/PET raw data section.
This section will be described more thoroughly in the MMVT addon documentation. For now, if you wish to render your data from the command prompt, you can run the following script:
python -m src.mmvt_addon.scripts.render_fmri_clusters -s subject_name -a atlas_name
The end result of this script is a split brain figure, with lateral and medial views:

This script will render the clusters for all the subject’s contrast you’ve calculated. If you are using colin27, that will render all the clusters for all the results loaded to colin27. For example, if you used SPM to calculate the difference between several subjects and a control group, the contrasts will be the subjects names, and the script will render the results of all the comparisons. Also, by default, it’ll render both the pial and half inflated brain and both in white and black backgrounds. For more details, take a look at the script’s documentation. If you are using a specific subject (not colin27), you’ll need to create the medial and lateral views manually, as described in the rendering panel documentation.
Given the low time resolution, only resting state data is supported for functional connectivity. To analyze the resting state fMRI data, you need to call the fMRI preprocessing as the following:
python -m src.preproc.fMRI -s subject-name -a atlas-name -f analyze_resting_state --fmri_file_template fmri-file-template
This call will load an fMRI file according to the given a template for each hemisphere, and calculate a time series for each label according to the given atlas (atlas-name). This function assumes that the data was already projected on the cortical surface. If this isn’t the case, you should first project it, by calling the fMRI preprocessing as described here. You just need to add the -f project_volume_to_surface flag to the call, to run only the projection function. For example, let’s say you’ve already processed your fMRI data with FsFast, and as result, you got two files, subject-name.siemens.sm6.fsaverage.rh.nii.gz and subjext-name.siemens.sm6.fsaverage.rh.nii.gz where subject-name is the name of your subject. First, you need to copy those files to the links/fMRI/subject-name/ folder. The fmri-file-template is {subject}.siemens.sm6.{morph_to_subject}.{hemi}.{format}. The code will put subject-name in {subject} and rh and lh in {hemi}. The default value for the format is ‘nii.gz’. In case you want to set a different format, you should use the –input_format flag. The {morph_to_subject} is used in case the data was projected to a template brain, fsaverage, like in the current example. In such a case, you’ll need to use the flag –morph_labels_to_subject. You can also use wildcards in the fmri-file-template. For example: “{subject}*{morph_to_subject}.{hemi}*.{format}”. That will work only if there is only one file matching this template. If your files are named ‘subject-name.siemens.sm6.fsaverage.rh.b0dc.nii.gz’ for example, this template will catch it. The default measure to calculate a time series for each label is just taking the mean of all the label’s vertices. The other option is ‘PCA’, where the first principal component is taken. To change this option you’ll need to use the –labels_extract_mode flag. The default atlas-name is aparc.DKTatlas40, as explained here. To summarize, in the current example the call is the following:
python -m src.preproc.fMRI -s subject-name -a atlas-name -f analyze_resting_state --fmri_file_template "{subject}\*{morph_to_subject}.{hemi}\*.{format}" --morph_labels_to_subject fsaverage
The resting state analysis function will create two files, one for each hemisphere: labels_data_{atlas-name}{labels-extract-mode}{hemi}.npz. The files are located in the links/mmvt_blend/subject-name/fmri folder. These npz files include two variables: data (labels_num x T), and names, the labels names.
If you wish to call this function in code, look at the function analyze_resting_state in src.preproc.examples.fMRI, or call it directly:
python -m src.preproc.examples.fMRI -s subject-name -a atlas-name -f analyze_resting_state
Remember that if you are using the aparc.DKTatlas40 atlas, you don’t need to use the -a flag.
If you don’t know the TR, you can user the FreeSurfer mri_info utility, or call this function:
python -m src.preproc.fMRI -s subject-name -f get_tr --fmri_fname fmri_fname
Where fmri_fname is your fMRI file’s name, like subject-name.fsaverage.rh.nii.gz for example.The only two supported file types are nii.gz and mgz. If fmri-fname type is nii.gz, the code will convert it to mgz.
The fMRI preprocessing creates two files, one for each hemisphere: labels_data_(atlas_name}{labels_extract_mode}{hemi}.npz. The files are located in the links/mmvt_blend/subject-name/fmri folder. These npz files include two variables: data (labels_num x T), and names, the labels names. To load them and run a functional connectivity analysis, you should call the connectivity preprocessing module as the following:
python -m src.preproc.connectivity -s subject-name -a atlas-name -f calc_lables_connectivity --connectivity_modality fmri --connectivity_method connectivity-method --windows_length windows-length --windows_shift windows-shift
Where the connectivity-method is the method you want to use to calculate the functional connectivity. The methods that are supported are Pearson correlation (corr) and Phase Lag Index (pli).The calculation will be done using sliding windows, according to the window-length (the length of the windows) and window-shift (the shift in each slide). You can also calculate the coefficient variation of the values by concatenating ‘cv’ (using a comma) to the connectivity-method. The unit of the length and shift of the windows is the fMRI TR. For example, if your fMRI TR is 3s, windows-length=20 and windows-shift=3, then the length of the windows will be 60s, and the windows shift 9s.
The end result is the following files in the links/mmvt_blend/subject-name/connectivity:
- fmri_connectivity-method.npy: The numpy matrix, NxNxW, where N is the number of the cortical labels and W is the number of windows.
- fmri_connectivity-method.npz: The connectivity properties for visualization.
- fmri_vertices.pkl: The vertices information for visualization.
In case you added cv to the connectivity-method, these files will also be created:
- fmri_connectivity-method_cv.npy: The coefficient variation of the fmri_connectivity-method matrix.
- fmri_connectivity-method_cv.npz: The coefficient variation connectivity properties for visualization.
- fmri_connectivity-method_cv_mean.npy: The mean of the fmri_connectivity-method_cv matrix.
- fmri_connectivity-method_cv_mean.csv: A cortical labels coloring file, which is saved in the ../coloring folder.
For example, if you wish to:
- Calculate the correlation between the fMRI BOLD signal of the cortical labels time course.
- Where the labels are parcellated according to the Lausanne125 atlas
- Calculate also the coefficient of variation for each label
- Use windows length of one minute and shift of 9s, where the TR is 3s
python -m src.preproc.fMRI -s subject-name -a laus125 -f calc_lables_connectivity --connectivity_modality fmri connectivity_method=corr,cv --windows_length 20 --windows_shift 3
If you wish to call this script from code, take a look at the example in src.preproc.examples.connectivity.calc_fmri_connectivity:
from src.preproc import connectivity as con
args = con.read_cmd_args(dict(
subject=args.subject,
atlas='laus125',
function='calc_lables_connectivity',
connectivity_modality='fmri',
connectivity_method='corr,cv',
windows_length=20,
windows_shift=3
))
con.call_main(args)
- Identifies stereotactic EEG (sEEG) electrodes and group them to different leads using a semi-automatic algorithm [48]. The electrodes coordinates are transformed from the CT scanner space to the subject’s MRI FreeSurfer RAS coordinates, tkreg RAS (Right-Anterior-Superior)
- Reads the European Data Format (EDF) raw data, highly used for EEG and invasive electrodes recordings in hospitals. The user can very easily set the start and end time of the exported EDF file to import into MMVT a window that captures the desired event (an epileptic seizure for example).
- Reads analyzed data saved in FieldTrip [9] format, a MATLAB toolbox for advanced analysis methods of MEG, EEG, and invasive electrophysiological data
- Transfers the electrodes coordinates to a template brain and between subjects using the FreeSurfer combined volumetric and surface registration (by wrapping the mri_cvs_register and apply_morph Freesurfer commands) [14], [49].
- Transforms electrodes from surface space to positions on the dural surface using a simulated annealing “snapping” algorithm which minimizes an objective energy function as in Dykstra et al. 2012 [18].
- Estimate the probability that a particular brain region contributes the source of the signal at each electrode by using the Electrodes Labeling Algorithm (ELA) [48]. It operates with the assumption that this probability is estimated based on the Euclidean distance between the electrode and the brain labels, given a cortical atlas.
- Calculates the frequency-domain and time-frequency-domain electrodes connectivity, based on estimates of the cross spectral densities and power spectral densities (CSD/PSD), by using the same approach as in the EEG/MEG connectivity analysis.
[1] B. Fischl et al., “Automatically Parcellating the Human Cerebral Cortex,” Cereb. Cortex, vol. 14, no. 1, pp. 11–22, Jan. 2004.
[2] A. M. Dale, B. Fischl, and M. I. Sereno, “Cortical surface-based analysis: I. Segmentation and surface reconstruction,” Neuroimage, vol. 9, no. 2, pp. 179–194, 1999.
[3] B. Fischl, M. I. Sereno, and A. M. Dale, “Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system,” Neuroimage, vol. 9, no. 2, pp. 195–207, 1999.
[4] B. Fischl, “FreeSurfer,” NeuroImage, vol. 62, no. 2, pp. 774–781, Aug. 2012.
[5] B. Fischl et al., “Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain,” Neuron, vol. 33, no. 3, pp. 341–355, Jan. 2002.
[6] A. Gramfort et al., “MNE software for processing MEG and EEG data,” NeuroImage, vol. 86, pp. 446–460, Feb. 2014.
[7] A. Gramfort et al., “MEG and EEG data analysis with MNE-Python,” Front. Neurosci., 2013.
[8] W. D. Penny, K. J. Friston, J. T. Ashburner, S. J. Kiebel, and T. E. Nichols, Statistical parametric mapping: the analysis of functional brain images. Elsevier, 2011.
[9] R. Oostenveld, P. Fries, E. Maris, and J.-M. Schoffelen, “FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data,” Comput. Intell. Neurosci., vol. 2011, p. 1, 2011.
[10] S. Pieper, M. Halle, and R. Kikinis, “3D Slicer,” in 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821), 2004, pp. 632-635 Vol. 1.
[11] F. Tadel, S. Baillet, J. C. Mosher, D. Pantazis, and R. M. Leahy, “Brainstorm: a user-friendly application for MEG/EEG analysis,” Comput. Intell. Neurosci., vol. 2011, p. 8, 2011.
[12] G. Niso et al., “MEG-BIDS, the brain imaging data structure extended to magnetoencephalography,” Sci. Data, vol. 5, Jun. 2018.
[13] C. J. Holmes, R. Hoge, L. Collins, R. Woods, A. W. Toga, and A. C. Evans, “Enhancement of MR images using registration for signal averaging,” J. Comput. Assist. Tomogr., vol. 22, no. 2, pp. 324–333, 1998.
[14] G. Postelnicu, L. Zollei, and B. Fischl, “Combined volumetric and surface registration,” IEEE Trans. Med. Imaging, vol. 28, no. 4, pp. 508–522, 2009.
[15] W. E. Lorensen and H. E. Cline, “Marching cubes: A high resolution 3D surface construction algorithm,” in ACM siggraph computer graphics, 1987, vol. 21, pp. 163–169.
[16] C. Liu, “An analysis of the current and future state of 3D facial animation techniques and systems,” PhD Thesis, School of Interactive Arts & Technology-Simon Fraser University, 2009.
[17] M. Schaer, M. B. Cuadra, L. Tamarit, F. Lazeyras, S. Eliez, and J.-P. Thiran, “A surface-based approach to quantify local cortical gyrification,” IEEE Trans. Med. Imaging, vol. 27, no. 2, pp. 161–170, 2008.
[18] A. R. Dykstra et al., “Individualized localization and cortical surface-based registration of intracranial electrodes,” NeuroImage, vol. 59, no. 4, pp. 3563–3570, Feb. 2012.
[19] R. S. Desikan et al., “An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest,” NeuroImage, vol. 31, no. 3, pp. 968–980, Jul. 2006.
[20] S. D. Roth, “Ray casting for modeling solids,” Comput. Graph. Image Process., vol. 18, no. 2, pp. 109–144, Feb. 1982.
[21] M. Reuter, H. D. Rosas, and B. Fischl, “Highly accurate inverse consistent registration: a robust approach,” Neuroimage, vol. 53, no. 4, pp. 1181–1196, 2010.
[22] P. Ablin, J.-F. Cardoso, and A. Gramfort, “Faster Independent Component Analysis by Preconditioning With Hessian Approximations,” IEEE Trans. Signal Process., vol. 66, no. 15, pp. 4040–4049, 2018.
[23] J. C. Mosher, R. M. Leahy, and P. S. Lewis, “EEG and MEG: forward solutions for inverse methods,” IEEE Trans. Biomed. Eng., vol. 46, no. 3, pp. 245–259, 1999.
[24] M. X. Huang, J. C. Mosher, and R. M. Leahy, “A sensor-weighted overlapping-sphere head model and exhaustive head model comparison for MEG,” Phys. Med. Biol., vol. 44, no. 2, p. 423, 1999.
[25] M. S. Hämäläinen and R. J. Ilmoniemi, “Interpreting magnetic fields of the brain: minimum norm estimates,” Med. Biol. Eng. Comput., vol. 32, no. 1, pp. 35–42, 1994.
[26] A. M. Dale et al., “Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity,” Neuron, vol. 26, no. 1, pp. 55–67, Apr. 2000.
[27] C. Piano et al., “Wake and sleep EEG in patients with Huntington disease: an eLORETA study and review of the literature,” Clin. EEG Neurosci., vol. 48, no. 1, pp. 60–71, 2017.
[28] B. D. Van Veen, W. Van Drongelen, M. Yuchtman, and A. Suzuki, “Localization of brain electrical activity via linearly constrained minimum variance spatial filtering,” IEEE Trans. Biomed. Eng., vol. 44, no. 9, pp. 867–880, 1997.
[29] K. Sekihara and S. S. Nagarajan, “Adaptive spatial filters for electromagnetic brain imaging,”, 2008.
[30] J. C. Mosher and R. M. Leahy, “Source localization using recursively applied and projected (RAP) MUSIC,” IEEE Trans. Signal Process., vol. 47, no. 2, pp. 332–340, 1999.
[31] J. C. Mosher and R. M. Leahy, “EEG and MEG source localization using recursively applied (RAP) MUSIC,” in Conference Record of The Thirtieth Asilomar Conference on Signals, Systems and Computers, 1996, pp. 1201–1207.
[32] J. Gro\s s, J. Kujala, M. Hämäläinen, L. Timmermann, A. Schnitzler, and R. Salmelin, “Dynamic imaging of coherent sources: studying neural interactions in the human brain,” Proc. Natl. Acad. Sci., vol. 98, no. 2, pp. 694–699, 2001.
[33] J. F. Hipp, A. K. Engel, and M. Siegel, “Oscillatory synchronization in large-scale cortical networks predicts perception,” Neuron, vol. 69, no. 2, pp. 387–396, 2011.
[34] M. van Vliet, M. Liljeström, S. Aro, R. Salmelin, and J. Kujala, “Analysis of functional connectivity and oscillatory power using DICS: from raw MEG data to group-level statistics in Python,” Front. Neurosci., vol. 12, 2018.
[35] B. Babadi and E. N. Brown, “A review of multitaper spectral analysis,” IEEE Trans. Biomed. Eng., vol. 61, no. 5, pp. 1555–1564, May 2014.
[36] D. J. Pearce, “An improved algorithm for finding the strongly connected components of a directed graph,” Vic. Univ. Wellingt. NZ Tech Rep, 2005.
[37] T. Zemen and C. F. Mecklenbrauker, “Time-variant channel estimation using discrete prolate spheroidal sequences,” IEEE Trans. Signal Process., vol. 53, no. 9, pp. 3597–3607, 2005.
[38] D. B. Percival and A. T. Walden, Spectral analysis for physical applications. cambridge university press, 1993.
[39] F. J. Harris, “On the use of windows for harmonic analysis with the discrete Fourier transform,” Proc. IEEE, vol. 66, no. 1, pp. 51–83, 1978.
[40] O. Rioul and P. Duhamel, “Fast algorithms for discrete and continuous wavelet transforms,” IEEE Trans. Inf. Theory, vol. 38, no. 2, pp. 569–586, 1992.
[41] C. Torrence and G. P. Compo, “A practical guide to wavelet analysis,” Bull. Am. Meteorol. Soc., vol. 79, no. 1, pp. 61–78, 1998.
[42] G. Nolte, O. Bai, L. Wheaton, Z. Mari, S. Vorbach, and M. Hallett, “Identifying true brain interaction from EEG data using the imaginary part of coherency,” Clin. Neurophysiol., vol. 115, no. 10, pp. 2292–2307, 2004.
[43] J.-P. Lachaux, E. Rodriguez, J. Martinerie, and F. J. Varela, “Measuring phase synchrony in brain signals,” Hum. Brain Mapp., vol. 8, no. 4, pp. 194–208, 1999.
[44] M. Vinck, M. van Wingerden, T. Womelsdorf, P. Fries, and C. M. Pennartz, “The pairwise phase consistency: a bias-free measure of rhythmic neuronal synchronization,” Neuroimage, vol. 51, no. 1, pp. 112–122, 2010.
[45] C. J. Stam, G. Nolte, and A. Daffertshofer, “Phase lag index: assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources,” Hum. Brain Mapp., vol. 28, no. 11, pp. 1178–1193, 2007.
[46] M. Vinck, R. Oostenveld, M. van Wingerden, F. Battaglia, and C. M. A. Pennartz, “An improved index of phase-synchronization for electrophysiological data in the presence of volume-conduction, noise and sample-size bias,” NeuroImage, vol. 55, no. 4, pp. 1548–1565, Apr. 2011.
[47] F. Pedregosa et al., scikit-learn: Machine Learning in Python’. 2010.
[48] N. Peled, T. Gholipour, A. Paulk, O. Felsenstein, and Dougherty DD, Widge AS, Eskandar EN, Cash SS, Hamalainen MS, Stufflebeam SM, “Invasive Electrodes Identification and Labeling,” github, 2017. .
[49] L. Zöllei, A. Stevens, K. Huber, S. Kakunoori, and B. Fischl, “Improved tractography alignment using combined volumetric and surface registration,” Neuroimage, vol. 51, no. 1, pp. 206–213, 2010.
